Share Important Moment of MileCell Bio with You
2025.04.25
The Mixed Lymphocyte Reaction (MLR), a gold-standard technique in transplant immunology, has emerged as a core functional platform for evaluating immune-modulatory effects in drug development. By simulating T-cell responses to allogeneic HLA class II antigens (HLA-DR/DQ/DP), MLR quantifies a drug’s ability to regulate T-cell activation, proliferation, and effector molecule secretion. This provides critical data for developing immunosuppressants, cell therapies, and bispecific antibodies (Morris et al., 2019)[1]. Compared to traditional in vitro models, MLR dynamically dissects a drug’s multidimensional impact on immune cascades, significantly reducing R&D risks from preclinical to clinical stages (CLSI I/LA31-A, 2014)[2].
MLR’s cascade offers multiple intervention nodes for drug development:
1. Antigen Recognition Blockade: Drugs targeting the TCR-pMHC II complex (e.g., HLA-blocking antibodies) inhibit naïve T-cell activation.
2. Costimulatory Signal Modulation: CD28/B7 or 4-1BB agonists enhance T-cell expansion efficiency.
3. Cytokine Pathway Suppression: JAK/STAT inhibitors block IL-2 signaling to suppress clonal proliferation.
4. Effector Phase Intervention: Granzyme inhibitors or Fas/FasL-neutralizing antibodies mitigate cytotoxicity (Macian et al., 2002)[3].
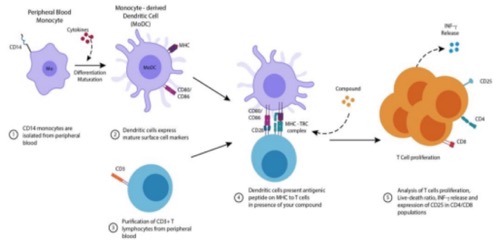
Figure 1 Mechanism of Mixed Lymphocyte Reaction (MLR) Illustrated
• JAK Inhibitors: In MLR models, Baricitinib (10 nM) reduced CD4+ T-cell proliferation by 62% (p<0.001) and IFN-γ secretion by 85%, directly supporting its approval for GVHD (Zeiser et al., 2021)[5].
• CTLA-4 Fusion Proteins: Abatacept competitively binds B7 molecules, lowering MLR stimulation indices (SI) from 5.3 to 1.8 (p=0.003), establishing it as a first-line treatment for rheumatoid arthritis (Buckner et al., 2019)[6].
• CAR-T Functional Validation: MLR assays showed that CD19 CAR-T cell killing of B lymphoblasts (CFSE decay rate) positively correlated with clinical response rates (r=0.79, p=0.01), outperforming traditional cytotoxicity assays (June et al., 2018)[7].
• Universal UCAR-T Screening: MLR identified TCR-knockout UCAR-T cells with 10-fold lower alloreactivity (SI<1.5) compared to conventional CAR-T (Depil et al., 2020)[8].
• CD3×CD20 Bispecific Antibodies: Glofitamab achieved 92% T-cell-mediated lymphoma cell killing in MLR, with controlled IL-6 release (<50 pg/mL), indicating clinical safety (Dickinson et al., 2022)[9].
• 4-1BB Agonists: Utomilumab (10 μg/mL) boosted CD8+ T-cell expansion by 3.2-fold (p=0.002) and promoted memory T-cell differentiation (Melero et al., 2023)[10].
• One-Way MLR: Mitomycin C-inactivated donor cells quantify drug effects on recipient T cells (³H-TdR incorporation CV<8%).
• Cytokine Storm Prediction: Luminex-based IL-6/IFN-γ peak monitoring predicts CRS risk (CLSI EP05-A3, 2014)[11].
• Dose-Response Modeling: MLR correlates IC50/EC50 with clinical effective doses (R²>0.9), accelerating IND submissions (Takebe et al., 2022)[4].
• Biomarker Discovery: Single-cell sequencing revealed Granzyme BhighCD8+ T cells (>15%) predict superior PD-1 inhibitor efficacy (Stubbington et al., 2017)[12].
• Microfluidic MLR Platforms (e.g., xCELLigence RTCA): Enable 96-hour dynamic monitoring of 200+ drug candidates (Li et al., 2021)[13].
• Liver Organoid + Allogeneic T Cells: Simulate tissue-specific immune modulation and hepatotoxicity risks (Takebe et al., 2022)[4].
• Machine Learning (e.g., Random Forest): Predicts clinical response probability using MLR data (proliferation rates, cytokine profiles, subset ratios; AUC=0.93) (Chen et al., 2023)[14].
From target discovery to clinical translation, MLR is evolving into a core decision-making tool for immune-modulatory drugs. By integrating automation, organoid models, and AI, MLR is transforming drug development from a “trial-and-error” approach to a “precision prediction” paradigm, accelerating safer and more effective therapies for patients worldwide.
[1] Morris GP et al. Front Immunol. 2019. DOI:10.3389/fimmu.2019.02349
[2] CLSI I/LA31-A. 2014.
[3] Macian F et al. Cell. 2002. DOI:10.1016/S0092-8674(02)00767-5
[4] Takebe T et al. Nat Biotechnol. 2022. DOI:10.1038/s41587-021-01085-1
[5] Zeiser R et al. Blood. 2021. DOI:10.1182/blood.2021011520
[6] Buckner JH et al. Nat Immunol. 2019. DOI:10.1038/s41590-019-0389-y
[7] June CH et al. Science. 2018. DOI:10.1126/science.aar6711
[8] Depil S et al. Nat Rev Drug Discov. 2020. DOI:10.1038/s41573-020-0071-2
[9] Dickinson MJ et al. NEJM. 2022. DOI:10.1056/NEJMoa2206913
[10] Melero I et al. Nat Rev Clin Oncol. 2023. DOI:10.1038/s41571-023-00746-1
[11] CLSI EP05-A3. 2014.
[12] Stubbington MJT et al. Nat Methods. 2017. DOI:10.1038/nmeth.4237
[13] Li Y et al. SLAS Discov. 2021. DOI:10.1177/24725552211006835
[14] Chen R et al. Nat Biotechnol. 2023. DOI:10.1038/s41587-023-01754-3